
Image via Mathagraphics/Shutterstock.com
The Double Anniversary of the Double Helix
It's been 60 years since DNA's discovery and 10 years since it was mapped.
Last week, April 25 marked a very special day. In 2003, Congress declared April 25 th DNA Day to mark the date that James Watson and Francis Crick published their seminal one-page paper in Nature describing the helical structure of DNA. That was 60 years ago. In that single page, they revealed how organisms elegantly store biological information and pass it from generation to generation; they discovered the molecular basis of evolution; and they effectively launched the era of modern biology.
But that’s not all that’s special about this date. It was ten years ago last month that we celebrated the completion of all of the original goals of the Human Genome Project (HGP), which produced a reference sequence of the 3 billion DNA letters that make up the instruction book for building and maintaining a human being. The $3 billion, 13-year project involved more than 2,000 scientists from six countries. As the scientist tasked with leading that effort, I remain immensely proud of the team. They worked tirelessly and creatively to do something once thought impossible, never worrying about who got the credit, and giving all of the data away immediately so that anyone who had a good idea about how to use it for human benefit could proceed immediately. Biology will never be the same. Medical research will never be the same.
We’ve discovered the 20,500 or so genes, and uncovered many of their regulatory switches, located in vast regions of DNA (once thought of as junk) that are vital for their proper function. We’ve catalogued genetic variations—some 54 million of them so far—that exist in the human species. Some of these variations are linked to disease—like heart disease, cancer, or diabetes—while others reveal specific adaptations that occurred as humans spread around the globe and adapted to different foods and climates. What’s more, we’ve discovered that our genome doesn’t work alone. Our bodies are continually interacting with microbes that are vital to our health, and we’re now decoding their genomes in what is called the microbiome project.
So how cheap is it to read your genome? Today, a scientist can decode it for just $3,000–$5,000 in only a few days; this continues to get cheaper and faster as technologies evolve. We have already done this for thousands of individuals (including my personal hero, South Africa’s Archbishop Desmond Tutu, who just won the Templeton Prize). We have discovered the genetic mutation behind almost 5,000 diseases or conditions. And we’ve harnessed this data to personalize medicine—we’ve matched more than 100 drugs to specific genes and developed genetic tests to help diagnose more than 1,000 diseases. Equally as important, we’re making sure that people can use these genetic tests without fear of discrimination. To this end, Congress passed the Genetic Information Nondiscrimination Act (GINA), a law banning genetic discrimination.
I am a physician; my dream is to see these advances in understanding the genome translated into better methods of prevention, treatment, and cure of disease. And we are seeing that happen in many areas—perhaps most prominently in cancer and in rare genetic diseases, as readers of this blog will have noted. But much work still lies ahead. As we celebrate this special day, let’s resolve to use all means possible to bring the promise of the genomic revolution to those billions of people in the world who are still waiting and hoping for its benefits to reach them.
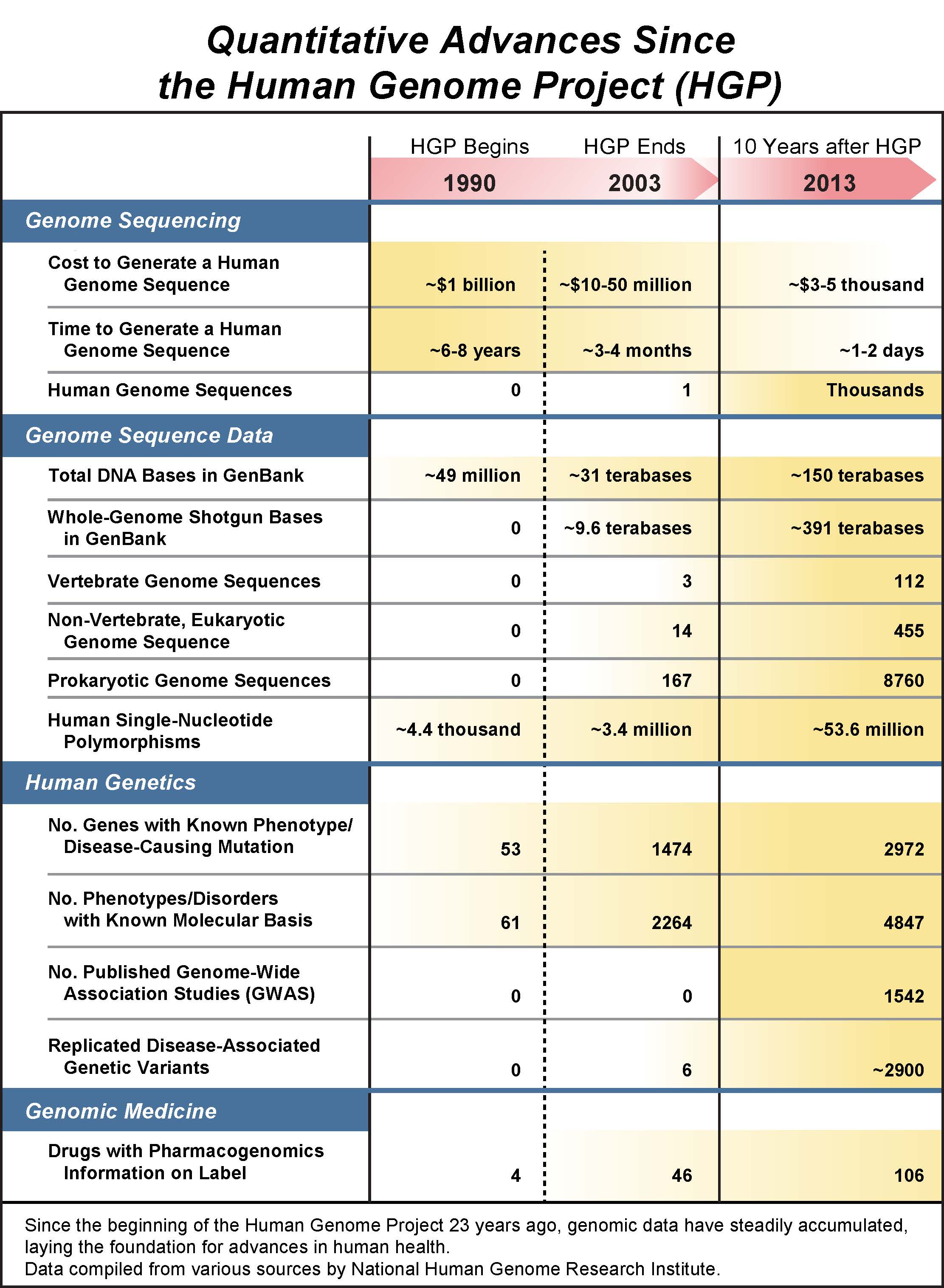
Image via Mathagraphics/Shutterstock.com







